RTX
A reasoning tool created for Translator that integrates many different databases into a cohesive knowledge graph.
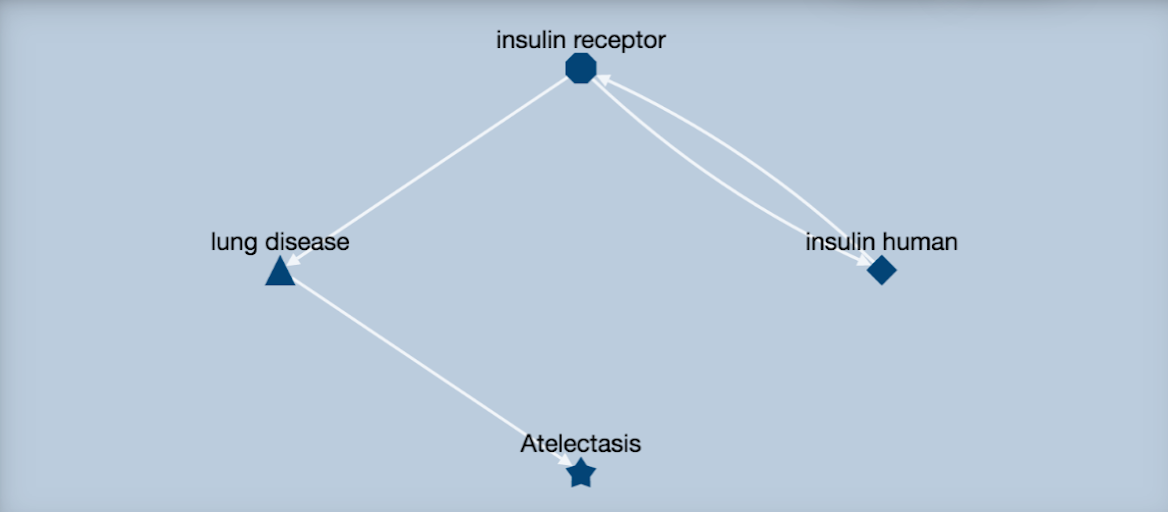
SemMedDB
A repository of semantic predications (subject-predicate-object triples) extracted by SemRep, a semantic interpreter of biomedical text. SemMedDB currently contains approximately 94.0 million predications extracted from PubMed.
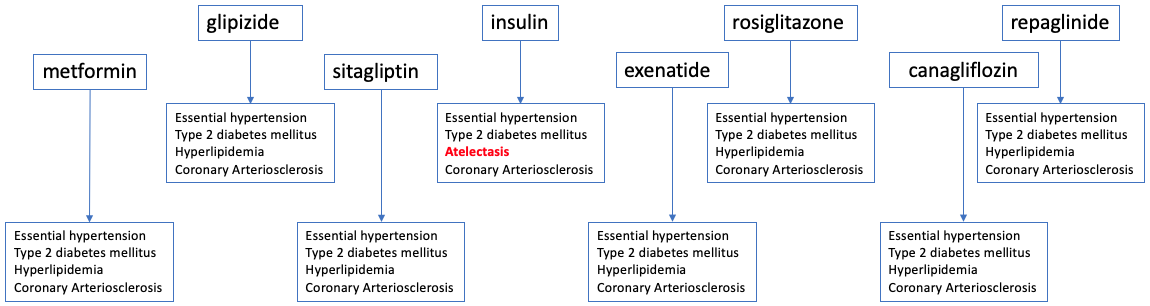
COHD
An open clinical database that includes counts and frequencies of conditions, procedures, drug exposures and patient demographics derived from the Columbia University Medical Center's Observational Health Data Sciences and Informatics (OHDSI) database.