Tiny Antibodies May Provide New Tool to Fight COVID-19
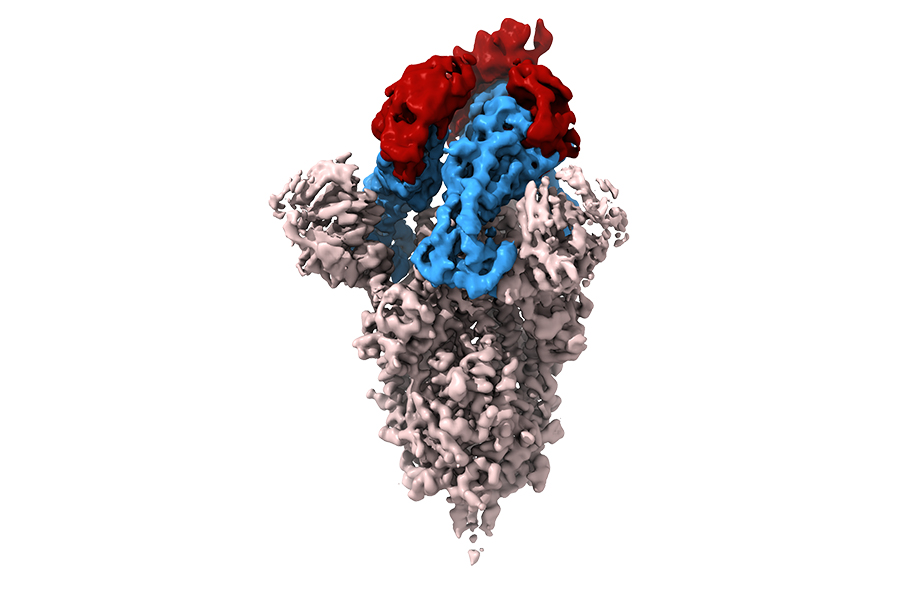
An NIH-led team of scientists built a library of small antibodies, called synthetic nanobodies, and used it to find promising new therapeutic leads for halting SARS-CoV-2 infection. The SARS-CoV-2 spike protein is depicted in white, and its three receptor binding domains (RBDs) are highlighted in blue. The RBD is the portion of the viral spike protein that binds to the protein receptor, ACE2, on the surface of healthy cells. SARS-CoV-2 enters cells through ACE2. Three nanobodies (red) mask the binding portion of the RBDs, preventing the spike protein from recognizing ACE2. This prevents the virus from entering the cell. (Kedar Sharma, Ph.D., and Mario Borgnia, Ph.D., National Institute of Environmental Health Sciences.)
August 30, 2022
As SARS-CoV-2 continues to evolve, scientists are on the hunt for therapeutics to combat new variants. To speed the search, an NIH-led team of scientists built a library of small antibodies, called synthetic nanobodies, and used it to find promising new therapeutic leads for halting viral activity.
The team from NCATS, the National Institute of Environmental Health Sciences and the Naval Research Laboratory reported their results in PLOS One.
Nanobodies are found in shark and camelid species, which include camels, llamas and alpacas. They are one-tenth the size of a human antibody. Their small size lets them attach inside protein grooves on viruses. This feature could help point scientists to weaknesses on the SARS CoV-2 spike protein that might otherwise be inaccessible to full-sized antibodies.
“Nanobodies make attractive building blocks for the design of new therapies,” said NCATS scientist Bryan Fleming, Ph.D., who helped lead the work, along with former NCATS scientist Ying Fu, Ph.D. “We’ve developed a way to rapidly and efficiently discover nanobodies against SARS-CoV-2.”
The first step to building the library was making llama nanobodies appear more like human antibodies. The researchers “humanized” the nanobodies by using a U.S. Food and Drug Administration–approved nanobody drug backbone as the basis for the library.
The researchers combined humanized “framework” regions, which act as support structures for nanobodies. These regions are shared among every nanobody in the library. Additionally, each nanobody has regions that recognize and stick to protein targets. These regions are like nanobody fingerprints and make each nanobody unique to a protein. Changing these regions creates new nanobodies capable of targeting new proteins.
According to Fleming, the construction of a synthetic library can yield about 10 billion possible nanobodies. These nanobodies can be studied as individual therapies or can be combined to create combination therapies.
“We hope that this approach for creating humanized nanobodies will accelerate the path to patients,” said NCATS translational scientist Matthew Hall, Ph.D., a co-author.
The scientists made several other different libraries to have a wider variety of nanobodies to choose from. They used NCATS’ high-throughput screening facilities to search through their nanobody libraries to quickly find those that bound tightly to and potentially would work best against the SARS-CoV-2 receptor-binding domain (RBD). The virus uses the RBD to dock with a cell and gain entry.
They identified three nanobodies that appeared to be most effective in blocking SARS-CoV-2 infection. The scientists tested the nanobodies against the B.1.1.7 (UK/alpha) SARS-CoV-2 variant, which was the dominant strain in the United States in early June 2020.
They also tested the three nanobodies on different modified viruses that can enter a cell but not cause disease. They found that high nanobody concentrations prevented infection, whereas low concentrations did not.
One of the nanobodies, RBD-1-2G, was especially strong in binding to the virus. The scientists modified RBD-1-2G, making different versions that were larger and closer to the structure of a more conventional antibody. They found that adding multiple RBD-1-2G nanobodies linked together into a single candidate therapeutic increased the effectiveness of blocking viral infection. A therapeutic composed of three nanobodies was found to be the most effective.
In tests against other variants, RBD-1-2G was very active in blocking the UK/alpha variant, but did not work well against variants, such as delta, mu and omicron. The scientists plan to continue to use the library to evaluate other nanobodies against SARS-CoV-2 variants and other viruses.
NCATS’ technology to develop a library of humanized nanobodies was among those NIH recently licensed to the Medicines Patent Pool through the World Health Organization COVID-19 Technology Access Pool. These licenses will enable manufacturers from around the world to use these technologies.
A nanobody library could prove useful for noninfectious diseases, as well.
“This is a huge library of antibodies that can be screened to identify drug candidates against almost any protein, related to almost any disease,” Hall said. “Given that there are over 10,000 rare diseases, most of which have no treatment, we hope this new platform will also prove useful in tackling that challenge.


