NCATS Researchers’ New Approach to Measure Gene Activity Could Speed Search for Rare Disease Therapies
March 23, 2021
NCATS scientists have designed an innovative test to find potential treatments for Charcot-Marie-Tooth disease (CMT), one of the most common forms of inherited neurological disease. Their new approach could speed the search for therapies for many more diseases where gene transcription is a therapeutic target.
CMT is a rare, hereditary disorder that damages the body’s nerves running from outside the brain and spine, leading to muscle weakness and loss. It affects about 126,000 people in the United States and 2.6 million people around the world. No cure exists, and current treatment primarily targets the disease’s symptoms, not its causes.
More than half of inherited cases are a subtype known as Charcot-Marie-Tooth 1A (CMT1A). In CMT1A, the body has too many copies of a gene for making peripheral myelin protein 22 (PMP22), a key protein used in the cells that form an insulating sheath around nerve fibers. Too much PMP22 can create a defective sheath that slows or stops a nerve fiber’s transmission of electrical signals, eventually causing muscle damage.
To help find potential new CMT1A therapies, NCATS biologist Natalia Martinez, Ph.D., and informatics engineer John Braisted collaborated with fellow NCATS translational scientists and researchers at the University of Wisconsin and the National Human Genome Research Institute. Their findings appeared online June 10 in ACS Pharmacology & Translational Science.
The researchers set out to create a test called an assay to identify compounds that could reduce PMP22 protein production. The assay needed to detect transcription, an important step in a cell’s protein-making process. But quickly detecting in thousands of samples whether a gene has been transcribed to ultimately make a protein has been a significant scientific challenge.
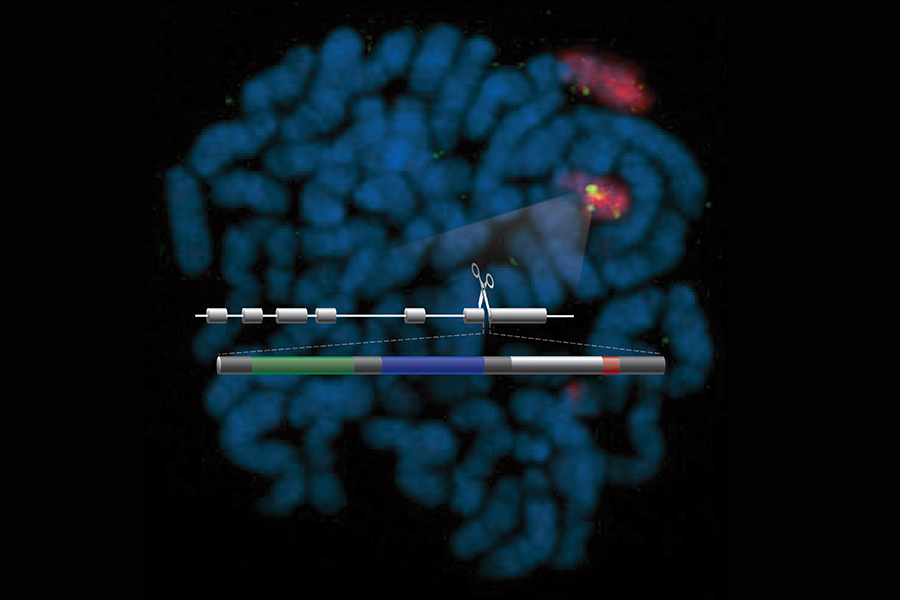
In earlier studies, the researchers devised a novel solution. They used gene editing technology to add the genetic code for a reporter enzyme into cells’ genes for PMP22. If the gene for the PMP22 protein is transcribed, the cell also will create the additional enzyme, which will report its presence by emitting a detectable bioluminescent glow.
There was a chance that drug compounds could interfere with the glowing reporter, however, which could lead to inaccurate assessments of a compound’s therapeutic potential. “Standard approaches to rapidly measure gene transcription in thousands of compounds can lead to large numbers of samples being identified as having positive results, when they are actually negative,” explained NCATS’ James Inglese, Ph.D., the study’s corresponding author.
So, in this latest study, the researchers added two different reporters into the gene for PMP22. Adding a second reporter — called the “coincidence reporter” approach — reduces the likelihood of an inaccurate transcription assessment, because the chance that a compound would interfere with both reporters is low.
In addition to detecting PMP22 gene transcription, the researchers needed to measure the transcription’s end product: PMP22 proteins. The scientists created a second assay. Once again, they turned to gene editing tools and a reporter that could show the PMP22 gene was transcribed and led to the creation of a protein.
With their two novel assays in hand, the researchers used quantitative high-throughput screening to rapidly test 42,577 compounds in NCATS’ chemical library to see how well they suppressed transcription of the PMP22 gene. The coincidence-reporter assay approach identified 181 compounds that reduced PMP22 transcription without harming cells. The researchers then used their second assay, which directly measured PMP22 protein levels, to find 16 compounds whose effect on transcription led to measurably lower PMP22 protein production.
The coincidence-reporter approach to measuring transcription activity is adaptable beyond the search for CMT1A treatments. “The coincidence reporter is a particularly effective tool in those diseases where a compound is needed that would suppress or activate a disease-related gene,” Inglese said.
For example, the technique could be tuned to search for therapeutic answers in hereditary neuropathy with liability to pressure palsies, a condition driven by too little PMP22. The researchers also have used coincidence reporters to find compounds with therapeutic promise for familial forms of Parkinson’s disease.