New Assay Technology Could Reshape Approach to Drug Discovery
June 3, 2025
NIH scientists have devised a relatively straightforward test based on the natural vibrations of proteins that could help improve the search for new drugs to more effectively target a wide range of diseases. The test, called the Structural Dynamics Response (SDR) assay, could address a key challenge in the hunt for new drugs: determining if and how well a drug candidate binds to a targeted protein.
The SDR assay lets drug developers quickly assess if and how strongly ligands — drug precursors that combine with target proteins — interact with a wide range of proteins. Scientists, led by James Inglese, Ph.D., at NIH’s NCATS, described their work in developing the assay in Nature Communications.
“The new technique promises to help speed — and simplify — drug discovery,” said Inglese, director of NCATS’ Preclinical Chemical Biology Laboratory. Turning a candidate molecule or drug into a treatment for patients can take more than a decade and hundreds of millions of dollars.
“This is close to a universal drug lead discovery and optimization platform for protein targets that are diverse in their structures and actions,” explained Inglese. “One of the most exciting aspects of this potentially transformative technology is that it can work without the need for target-specific reagents or specialized instruments.”
In laboratory experiments, Inglese and his NCATS colleagues tested SDR on target proteins from different classes, including kinases. Kinases play many important roles in cells, including in communication and growth, often making them potential targets for drugs. The researchers wanted to see if SDR could spot known inhibitors and potential protein ligands.
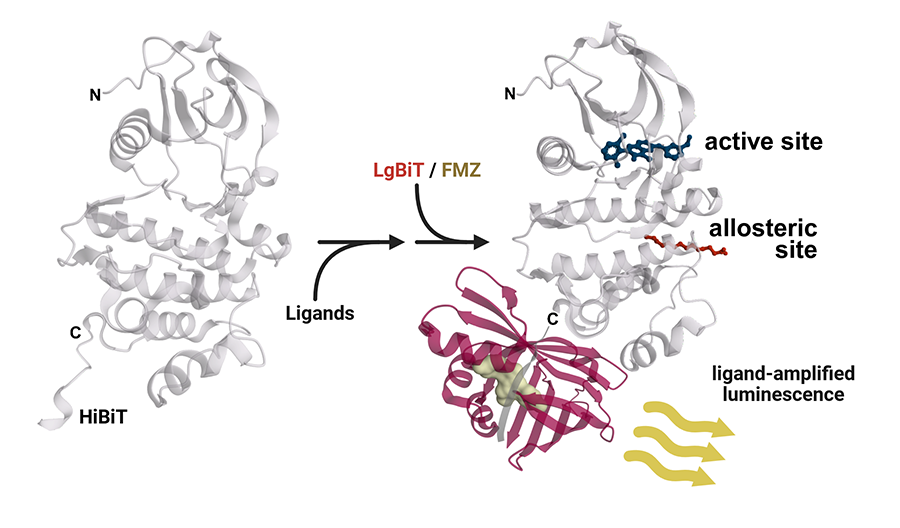
In one experiment, researchers compared SDR to a standard kinase enzyme assay across a collection of 128 compounds that target kinases, including the Abelson tyrosine kinase (ABL1). ABL1 mutations can affect cell growth and play roles in cancer. The scientists learned that SDR found ABL1 inhibitors as well as or better than the enzyme assay. The SDR assay also could detect compounds binding at a remote allosteric site on ABL1, which the standard kinase activity assay didn’t find. An allosteric binding site is an alternate location on a target protein that’s different from the normal “active” site.
The assay’s simplicity is key, Inglese noted. The detection of allosteric compounds showed that SDR doesn’t require users to measure proteins’ functions or even know what they do. In addition, the amount of a protein SDR needs is just a fraction of what standard tests need. And unlike standard methods that typically work only for a specific type of protein, SDR works across a wide range of proteins.
“SDR requires no specialized techniques, substrates, cofactors or other problematic materials,” said NCATS postdoctoral fellow and lead author Daniel Ciulla, Ph.D.
That’s especially important when a target protein has no known substrate to use in a test, added NCATS study co-author Patricia Dranchak, Ph.D. “We can eliminate the need for substrates and just measure direct binding to proteins,” she explained. A substrate is a substance that is acted on and changed.
SDR uses the natural motion and vibration of proteins. Ligand binding often changes the structure of proteins. This can range from rearranging protein geometry to subtly lowering the vibrations of protein motions. Such changes underlie the function of proteins in living systems, such as speeding chemical reactions or sending cell signals.
SDR measures the change between the motion of a protein’s ligand-free and its ligand-bound states by altering the light output of a sensor protein. The NCATS team found that the enzyme NanoLuc luciferase (NLuc) makes an ideal sensor protein because the intensity of light it gives off is readily modulated by the attached target protein’s ligand-influenced motions.
The light output of NLuc can be further increased using a “split” version of the sensor protein. In this case, scientists attach a small piece of NLuc to the target protein. When the NLuc is reformed by adding back the missing larger NLuc fragment, SDR is measured as a change in light intensity generated from the intact sensor protein.
By observing the amount of light made by a ligand-protein complex and the resulting rebuilt sensor protein, researchers can determine whether and how strongly a drug ligand interacts with a target protein. NCATS scientific director Matthew Hall, Ph.D., likens the approach to how a finger tap changes the glow of a touch-sensitive light bulb.
A technique called quantitative high-throughput screening (qHTS) lets scientists quickly test thousands of drug molecules at once in different dosages. Unproven newly discovered target proteins don’t always work with qHTS or any high-throughput screening method without extra steps. Inglese noted that SDR could help scientists find and focus on promising drug compounds faster. “SDR could be valuable throughout the drug development process, ranging from discovering new drug candidates to finding the best versions of the most promising compounds,” he said.
Related Content
Assay Development and Screening Technology (ADST)
Our scientists explore and develop new ways to quickly test and screen potential drug candidates, often in collaboration with disease foundations.
Our Impact on Tools and Technology
We create solutions that work for many diseases, speeding more treatments for all people more quickly.