Collaboration
We use powerful gene perturbation tools — such as CRISPR, RNAi and small molecule screening — to investigate how genes function, contribute to disease and can be targeted to develop better treatments.
Collaboration
At the Functional Genomics Laboratory, we collaborate with other research laboratories on a variety projects. We believe that these partnerships are essential for advancing our expertise in the field. By exploring different assay methodologies, we aim to further understand complex biological systems and tackle a wide range of diseases.
In the realm of gene perturbation technologies, we frequently use arrayed and pooled RNAi and CRISPR screens. Both RNAi and CRISPR knockout/interference (CRISPRi) techniques are utilized to interrogate gene function by blocking gene expression at different stages; CRISPR activation (CRISPRa) is employed to enhance gene expression.
FGL scientists also conduct arrayed chemogenomic screens using NCATS' annotated compound libraries. These include the NCATS Pharmaceutical Collection (NPC) approved drug library, the NCATS Pharmacologically Active Chemical Toolbox (NPACT), and the NCATS Mechanism Interrogation PlatE (MIPE) libraries. Our assay readouts vary depending on the specific assay and the biological context. Readouts range from straightforward metrics, like cell viability, to more complex evaluations, such as high-content imaging and next generation sequencing (NGS).
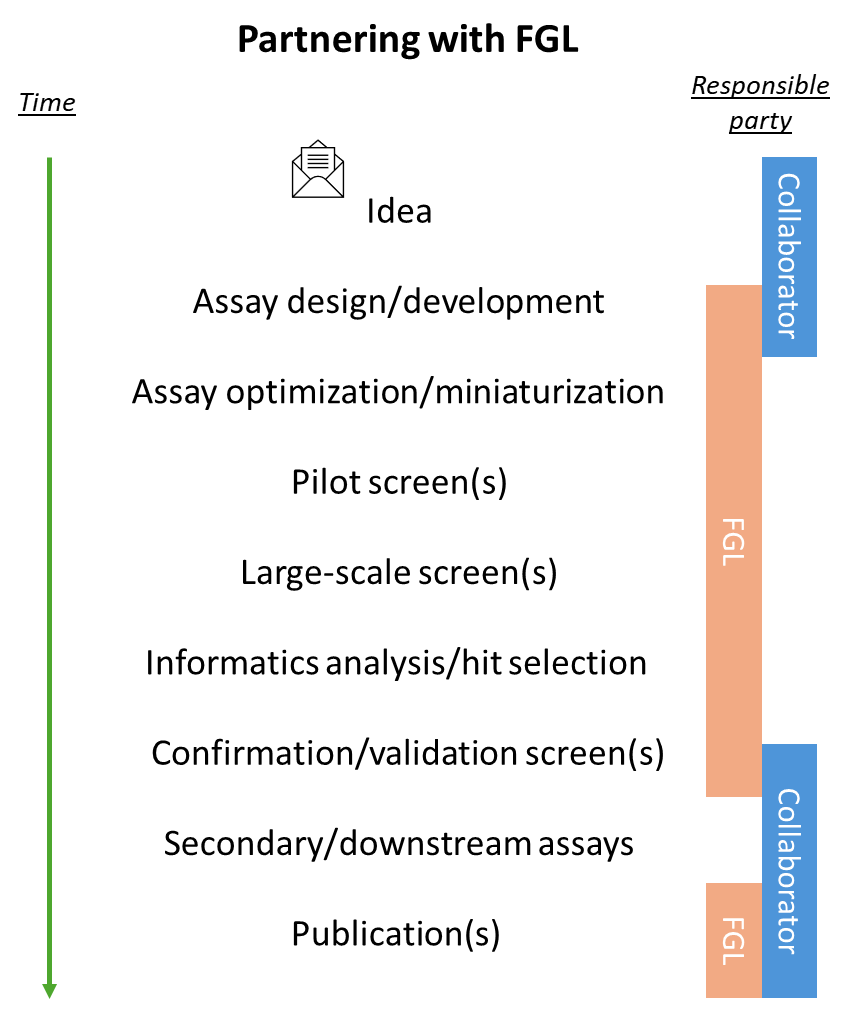
In a dynamic, collaborative process, FGL staff work with researchers throughout the planning phases as well as the execution of each research project. They do so by offering insight and technical assistance on 1) assay design and development, 2) screening, 3) bioinformatic analysis and 4) follow-up mechanistic studies.
FGL members are dedicated to training the next generation scientists interested in translational research. We welcome visiting scientists from our collaborative laboratories, providing them with opportunities to enhance their skills in functional genomic screening while working on the projects under the mentorship of FGL staff.
High-throughput siRNA screen for modulators of the nonsense-mediated mRNA decay (NMD) pathway
High-throughput siRNA screen for modulators of the nonsense-mediated mRNA decay (NMD) pathway
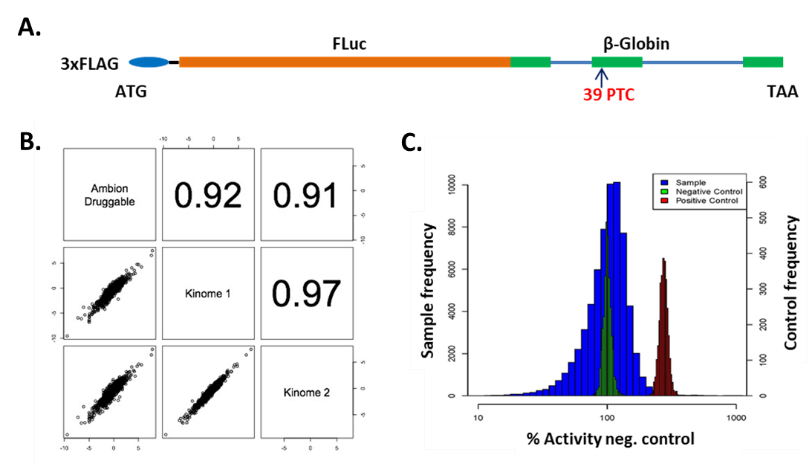
- Schematic of the NMD reporter construct (3xFLAG-FLuc-ß- globin^39UGA), in which a premature termination codon (PTC) in the second exon of the ß-globin gene triggers NMD.
- Demonstration of screen quality. HEK293 cells stably expressing the NMD reporter were screened with the Ambion kinome library (biological replicates) and subsequently with the druggable genome library. Strong correlations in reporter activity among replicate kinase siRNAs highlight the reproducibility and quality of the assay.
- Reporter activity distribution for positive (siUPF1) and negative controls (siNeg), alongside genome-wide siRNAs, further validate the robustness of the screen. Jolly et al., eLife, 2018.
This project was conducted in collaboration with Dr. J. Robert Hogg (NHLBI).
High-throughput siRNA screen for modulators of the nonsense-mediated mRNA decay (NMD) pathway

- Schematic of the NMD reporter construct (3xFLAG-FLuc-ß- globin^39UGA), in which a premature termination codon (PTC) in the second exon of the ß-globin gene triggers NMD.
- Demonstration of screen quality. HEK293 cells stably expressing the NMD reporter were screened with the Ambion kinome library (biological replicates) and subsequently with the druggable genome library. Strong correlations in reporter activity among replicate kinase siRNAs highlight the reproducibility and quality of the assay.
- Reporter activity distribution for positive (siUPF1) and negative controls (siNeg), alongside genome-wide siRNAs, further validate the robustness of the screen. Jolly et al., eLife, 2018.
This project was conducted in collaboration with Dr. J. Robert Hogg (NHLBI).
Resources
We foster collaboration to offer a wide range of screening libraries and instruments tailored to the unique needs of each project. Our in-house libraries are regularly updated to include the latest advancements in the field. We also welcome the opportunity to co-develop custom libraries to meet specific project requirements. Below are examples of our in-house libraries.
Arrayed 384-well CRISPR/Cas9 Knockout (KO) library:
- EditCo/Synthego Human sgRNA Library: whole-genome and pathway-specific; three sgRNAs/gene
Various pooled CRISPR KO, CRISPRi, and CRISPRa lentiviral libraries
Arrayed 384-well RNAi library:
- Human Dharmacon ON-TARGETplus siRNA Library: whole-genome and pathway-specific; set of four siRNAs
- Mouse Dharmacon ON-TARGETplus siRNA Library: druggable and pathway-specific; set of four siRNAs
- Pathway-focused and custom siRNA libraries are also available, tailored to specific project needs.
Arrayed 1536- and 384-well small molecule chemogenomic library, quantitative high-throughput screening (qHTS) format:
- NCATS Pharmaceutical Collection (NPC): FDA-approved and investigational drugs
- Mechanism Interrogation PlatEs (MIPE): Compounds annotated by mechanism of action
- NCATS Pharmacologically Active Chemical Toolbox (NPACT): Tool compounds targeting a wide range of biological pathways
- Additional targeted and custom libraries: Small molecules focused on specific pathways or target classes such as kinases and epigenetic modulators
For additional information about NCATS’ small molecule libraries, visit the Compound Management page.
Additional libraries, such as cDNA and small peptide libraries, are available and can be tailored to specific needs.
High-content, high-throughput (1,536-well) assay to identify compounds modulating the autophagy pathway
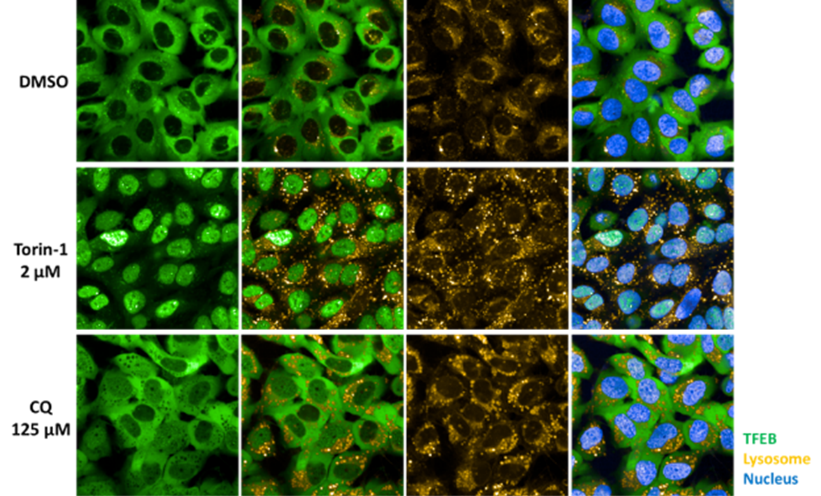
High-content, high-throughput (1,536-well) assay to identify compounds modulating the autophagy pathway
U2OS cells stably expressing GFP-TFEB were treated for 6 hours with DMSO, Torin-1, or Chloroquine (CQ), then stained with LysoTracker (yellow) and DAPI (blue). Nuclear translocation of GFP-TFEB was observed in Torin-1 and CQ-treated cells, with enlarged lysosomes visible in CQ-treated cells.
High-content, high-throughput (1,536-well) assay to identify compounds modulating the autophagy pathway

U2OS cells stably expressing GFP-TFEB were treated for 6 hours with DMSO, Torin-1, or Chloroquine (CQ), then stained with LysoTracker (yellow) and DAPI (blue). Nuclear translocation of GFP-TFEB was observed in Torin-1 and CQ-treated cells, with enlarged lysosomes visible in CQ-treated cells.
Bioinformatics
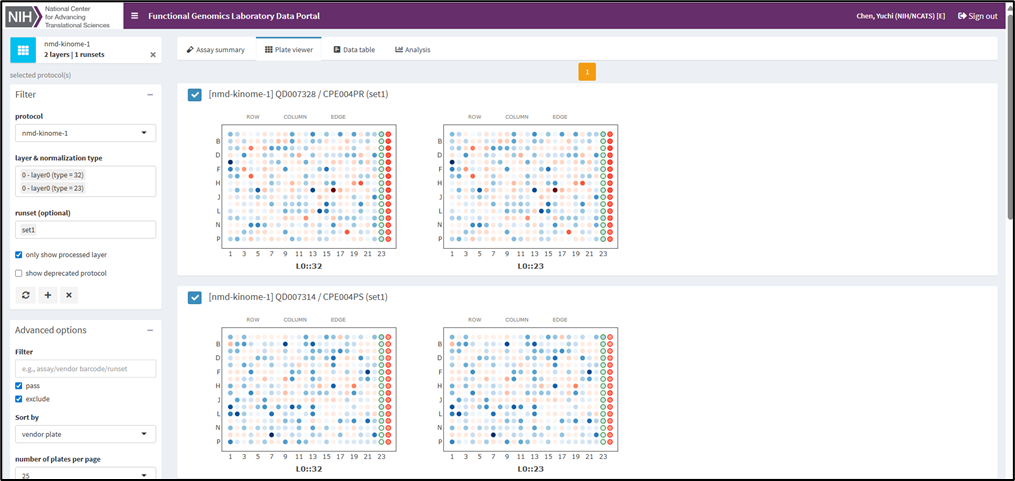
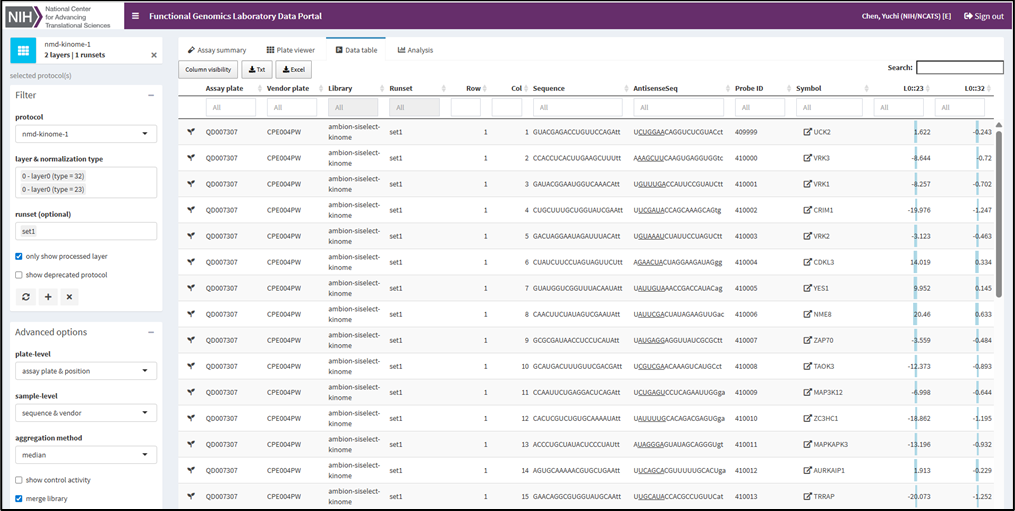
FGL Bioinformatician, Ryan L. MacArthur, Ph.D., specializes in analyzing data from high-throughput screening. The team uses a custom data client to visualize screening results. This streamlines quality control and hit selection, and it enhances the overall efficiency of our research.
Please contact Ken Cheng, Ph.D., about collaborating with the Functional Genomics Laboratory.